What's The Science?
Background
Coronavirus Disease (COVID-19), a lower respiratory tract pneumonia caused by the novel coronavirus SARS-CoV2, emerged as a global pandemic in late 2019. UConn, like most universities across the US, has been faced with the challenge of containing and mitigating the spread of the highly infective SARS-CoV2 virus so that the university can remain open without compromising the safety of our students, staff and faculty.
A broad surveillance approach that employs multiple modalities that is fast, affordable and scalable is critically needed to effectively screen members of the campus community for SARS-CoV2, support targeted responses to emerging clusters of cases, and contain and mitigate COVID-19, and thus prevent outbreaks. We have assembled a team among campus community partners to develop an integrated surveillance approach using existing expertise and adapting cutting edge molecular and data visualization technologies for our university.
Overview
Pooled Surveillance Testing
Our pooled surveillance testing approach relies on two different sources of testing material: wastewater to detect SARS-COV2 in feces and a gargle lavage to detect SARS-COV2 in the respiratory system. Both types of samples are subjected to the same test: qRT-PCR, the standard laboratory method to measure viral load for RNA viruses (targeting two genes present in SARS-CoV2: E and N).
SARS-COV2: The SARS-CoV-2 virus has a positive-sense RNA genome ~29.9 kilobases that contains as many as 29 open reading frames. There are at least 16 nonstructural proteins, four structural proteins, and at least six or seven accessory proteins, although the exact number of total proteins coded by the viral genome is currently unknown. The two genes targeted in our qRT-PCR assay are based on the CDC target (N) and the WHO target (E), both of which are in the ORF2-ORF10 region. The function of the nucleocapsid, coded by the N gene, is to bind viral genomic RNA and form a ribonucleocapsid involved in genome protection, replication, assembly and immune evasion. The envelope small membrane protein, coded by the E gene, is a single-pass type III membrane protein involved in viral assembly, budding, and pathogenesis.
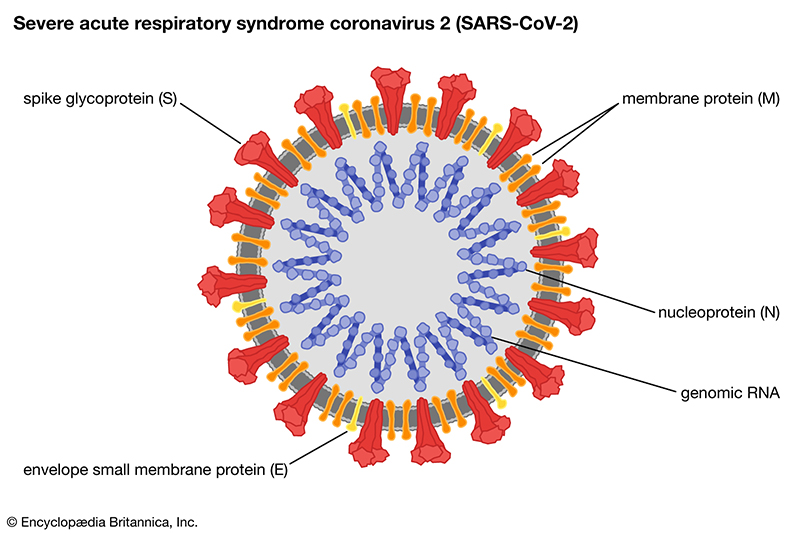
By targeting both the N and E genes in one reaction, we will most likely detect the presence of the SARS-COV2 virus even if there are unexpected nucleotide changes in the targets as drift of both sequences in the same sample is unlikely. We perform each qRT-PCR with a control gene to detect other RNAs and thus ensure a negative is reported for a lack of SARS-COV2, not for low sample integrity. For gargle samples, we use RNAse P for the detection of human nucleic acids. For wastewater, we use the pepper mild mottle virus, PMMoV, that is excreted from humans in high concentrations due to the consumption of foods that contain infected peppers.
SARS-CoV-2 genome

Wastewater
A composite sampler is placed in a manhole targeting a specific sewershed on campus. Daily collections from this pump are processed by concentrating viruses using nanoparticles and extracting RNA, followed by qRT-PCR. To date, 16 pumps have been placed on campus and samples are collected and tested daily.
Geographic Information Systems (GIS) mapping has allowed the team to identify the buildings producing the wastewater, thus delineating the presence of SARS-CoV2 in specific areas on campus (e.g. central wastewater processing plant, dormitory complexes, etc). Notably, ours is the first group to use nanoparticle concentration for wastewater samples which allows a reduced concentration rate of 12 samples per hour (versus the 12 samples in nine-hours typical of the standard filtration method). A small amount of extracted RNA is then placed in a multiplex qRT-PCR, containing probes and primer cocktails for RNAseP, N, and E.

Gargle Lavage
Upon receipt of gargle lavage sample collection tubes, we heat the tubes to inactivate the virus. Following inactivation, we combine up to 10 samples into one tube and place 100 uL of this mixture in a 96 well plate containing proteinase K. Proteinase K is an enzyme that, when heated, will function to break down or digest cellular proteins so that in later steps, the viral RNA, if present, will be accessible for reverse transcription and PCR using targeted primers and probe combinations. A small amount of the proteinase k-treated mixture is then used as a template for copying in a real-time quantitative RT-PCR for multiple probes and primer cocktails; RNAseP, N, and E. This molecular test is called multiplex qRT-PCR and allows for the determination of viral gene copy numbers in the pooled samples.